ebFRET
ebFRET performs combined analysis on multiple single-molecule FRET time series to learn a set of rates and states
Consensus analysis of many single-molecule FRET time series
ebFRET is a library written in Matlab for the analysis of single-molecule FRET time series with hidden Markov models.
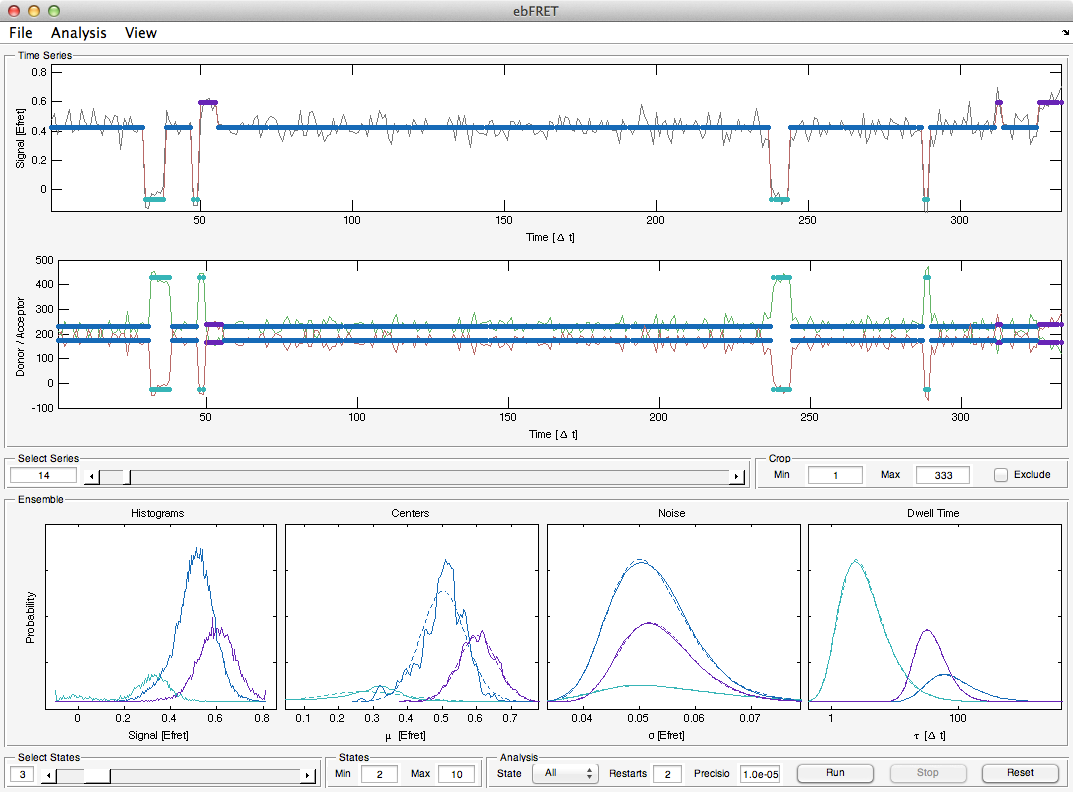
ebFRET is the successor of vbFRET, which it improves upon by performing combined analysis of multiple time series at once. This approach, known as empirical Bayes, is both more accurate and statistically robust. It also enables more advanced analysis use cases such as sub-population detection.
Get the Source
The ebFRET source code (with graphical user interface) can be downloaded here [git; zip]. Please see README and User Guide for installation instructions.
A command-line version of the code is also available for advanced analysis tasks, such as detection of subpopulations, which cannot be directly accomplished in the GUI [git; zip].
Code to plot transition density plots (TDP) as per “Observation and Analysis of RAD51 Nucleation Dynamics at Single-Monomer Resolution” in Methods in Enzymology by Subramanyam et al. can be found here: [git].
Papers
“Hierarchically-coupled hidden Markov models for learning kinetic rates from single-molecule data.” Jan-Willem van de Meent (@jwvdm), Jonathan E. Bronson, Frank Wood, Ruben L. Gonzalez Jr., Chris H. Wiggins. International Conference on Machine Learning 2013 [pdf]
“Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments.” Jan-Willem van de Meent (@jwvdm), Jonathan E. Bronson, Chris H. Wiggins, Chris H. Wiggins, Ruben L. Gonzalez Jr. Accepted for publication in the Biophysical Journal [pre-print; supplementary]
Slides
“Learning Kinetic Models from Single-Molecule Experiments”, ICML 2013 [pdf]
“Learning Kinetic Pathways from Single-Molecule FRET Measurements”, Columbia 2012 [pdf]